Thermodynamics and kinetics of protein-protein interactions for proteins involved in protein ubiquitination
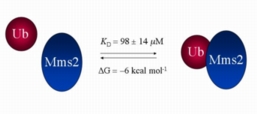
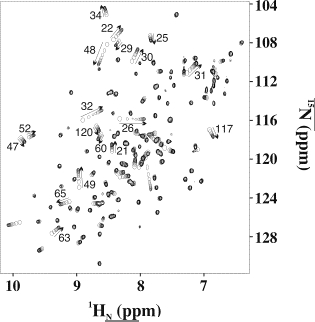 The figure on the right shows a superposition of 1H-15N NMR spectra of the protein Mms2 upon successive additions of the protein ubiquitin. The backbone amide cross peaks whose chemical shifts change give an the approximate location of the ubiquitin binding site on Mms2. In addition, we can plot the chemical shift changes as a function of the ligand:protein ratio to determine a dissociation constant.
The figure on the right shows a superposition of 1H-15N NMR spectra of the protein Mms2 upon successive additions of the protein ubiquitin. The backbone amide cross peaks whose chemical shifts change give an the approximate location of the ubiquitin binding site on Mms2. In addition, we can plot the chemical shift changes as a function of the ligand:protein ratio to determine a dissociation constant.
New NMR methods for thermodynamics and kinetics of protein-ligand interactions
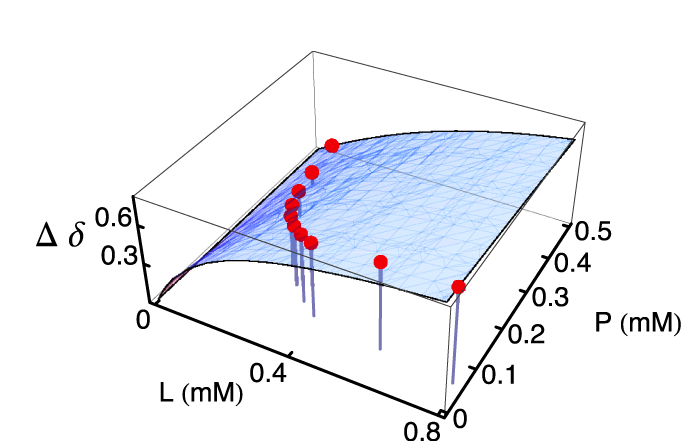
NMR is an ideal method for determining the dissociation constants KD of weak protein-ligand interactions. For a typical NMR titration, the initial protein concentration is held nearly constant, and for this condition, the protein concentration needs to be about half the dissociation constant to determine an accurate KD value, and the binding site needs to be saturated with ligand at the end of the titration, this requires a lot of ligand. We developed methods where co-variation of the protein and ligand concentrations gives more precise KD values, and accurate and precise kinetics.