Structures of proteins involved in protein ubiquitination
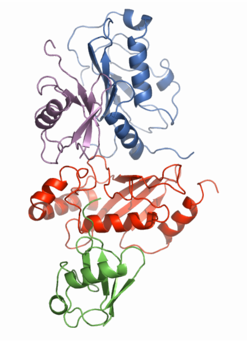
One of the main research goals of the laboratory is to determine the mechanism of lysine-63 linked protein ubiquitination using solution NMR. The picture on the right shows an NMR-based model for the Mms2-Ubc13-Ub2 complex. Given the large size of this protein complex, traditional solution NMR approaches are not suitable for structure determination. We were able to develop this structural model by assembling various protein components, and determining chemical shift changes in the NMR spectra upon mixing various protein partners. Combining chemical shift changes with the crystallographically determined structures of ubiquitin and the Mms2-Ubc13 heterodimer gives us a rough idea where the binding interfaces are for the various proteins.
We have also focused our research efforts on the interaction of Mms2 with ubiquitin (the marine and violet proteins in the figure to the right and the protein complex on the main home page). For this structure, we combined crystallographically determined structures and proton-proton NOEs, proportional to the distance between protons, to develop a higher resolution model for this protein-protein interaction.
Protein Structures in the RCSB Protein Data Bank
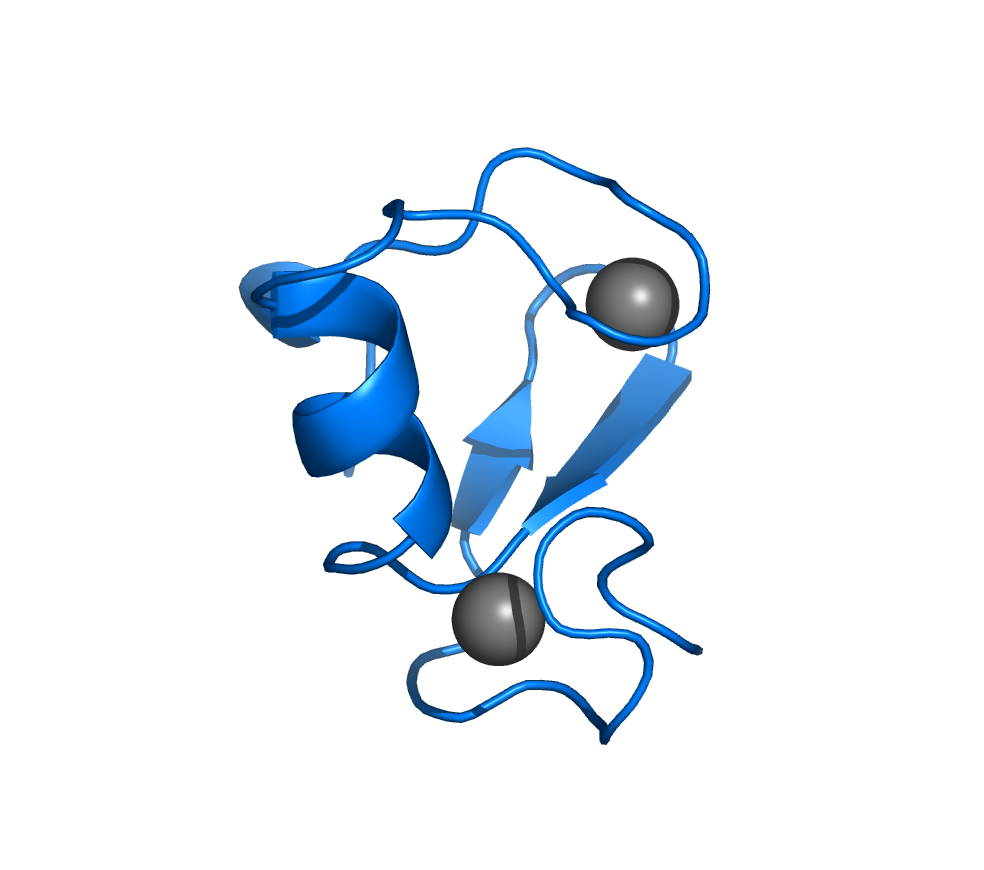
- RING domain from human TRAF6: 2JMD.
- Ubiquitin-conjugating variant human Uev1a: 2HLW.
- Human Mms2-ubiquitin complex: 1ZGU.
- Tandem ubiquitin interacting motifs (UIMs) from RAP80: 2MKG.
- Tandem UIMs from the ΔE81 deletion mutant from RAP80: 2MKF.
- Phosphorylated SIM from RAP80 bound to SUMO2: 2N9E.
NMR Spectra
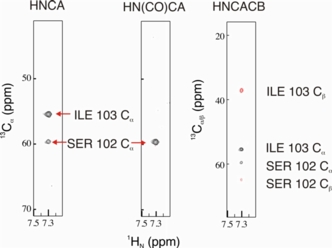
The NMR spectra on the right allowed for sequence specific backbone assignment of the 26 kDa Mms2-ubiquitin protein complex. These types of spectra are usually acquired for high molecular weight proteins that are isotopically enriched with 15N, 13C, and 2H.
- The HNCA correlates the 1HN-15N pair of residue i with the 13Cα of residues i and i-1.
- The HN(CO)CA is used with the HNCA and establishes the interresidue correlation for the 1HN-15N pair of residue i with the 13Cα of residues i-1.
- The HNCACB correlates the 1HN-15N pair of residue i with the 13Cα and 13Cβ of residues i and i-1.