NMR relaxation measurements of proteins involved in protein ubiquitination
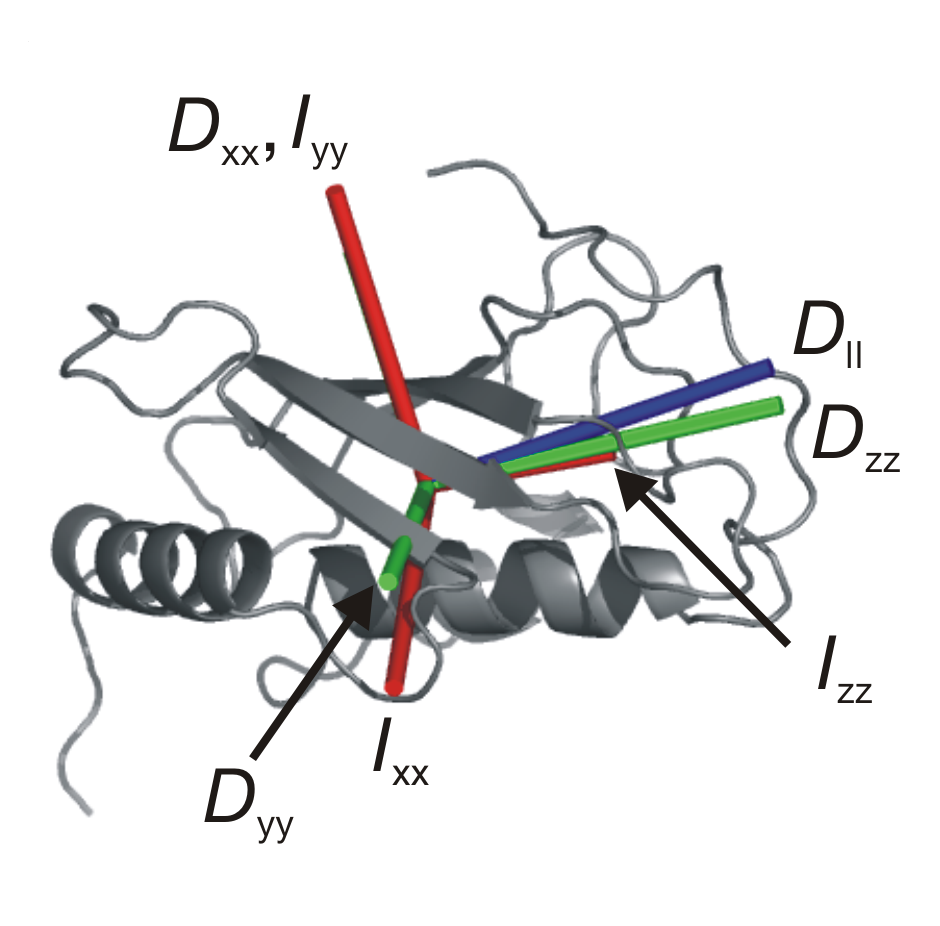
A detailed view of protein function requires high resolution atomic structures of proteins, as well as information regarding the rates and amplitudes of motions for the protein atoms. Solution state 15N relaxation measurements provide a powerful approach to characterizing the overall molecular tumbling of proteins, as well as their internal motions. The picture on the right shows the hydrodynamic properties of the protein Mms2. The shape of a protein molecule determines the nature of its rotational diffusion in solution. The main axes of rotational diffusion measured by NMR (in blue) match the expected main axes (in green), as well as the main axes of inertia (in red).
Main chain amide dynamics
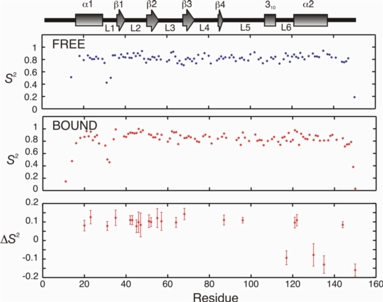
Main chain 15N relaxation data can be analyzed to yield an order parameter, or S2 value on a per residue basis. The magnitude of S2 varies from 0 (no motional restriction) to 1 (complete motional restriction). The figure on the right shows changes in S2 (bound - free) for Mms2 upon ubiquitin binding. Changes in S2 can be used to calculate semi-quantitative changes in entropy for the main chain.
Methyl side chain dynamics
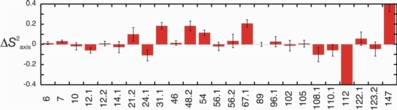
Much like 15N relaxation data for the protein main chain, methyl side chain 2H relaxation data can be analyzed to yield a methyl axis order parameter, or S2axis value on a per residue basis. The figure on the right shows changes in S2axis (bound - free) for Mms2 upon ubiquitin binding.